Carrot’s Data Engineering Procedure for OMOP
Carrot’s Data Engineering Process for OMOP standard vocabulary process primarily consist of Data Pre-processing, Metadata Profiling, Metadata Mapping, and Data Transformation.
The following documentation sets a Standardised Operating Procedure (SOP) that can be followed and applied during the engineering of data for conversion to an OMOP representation.
Figure 1 illustrates the 3 main data engineering stages and the associated software tools that are used, this process is commonly known as an Extract, Transform and Load (ETL) process.
This document only covers the ‘Extract and Transform’ sections of an ETL process as per the Figure below.
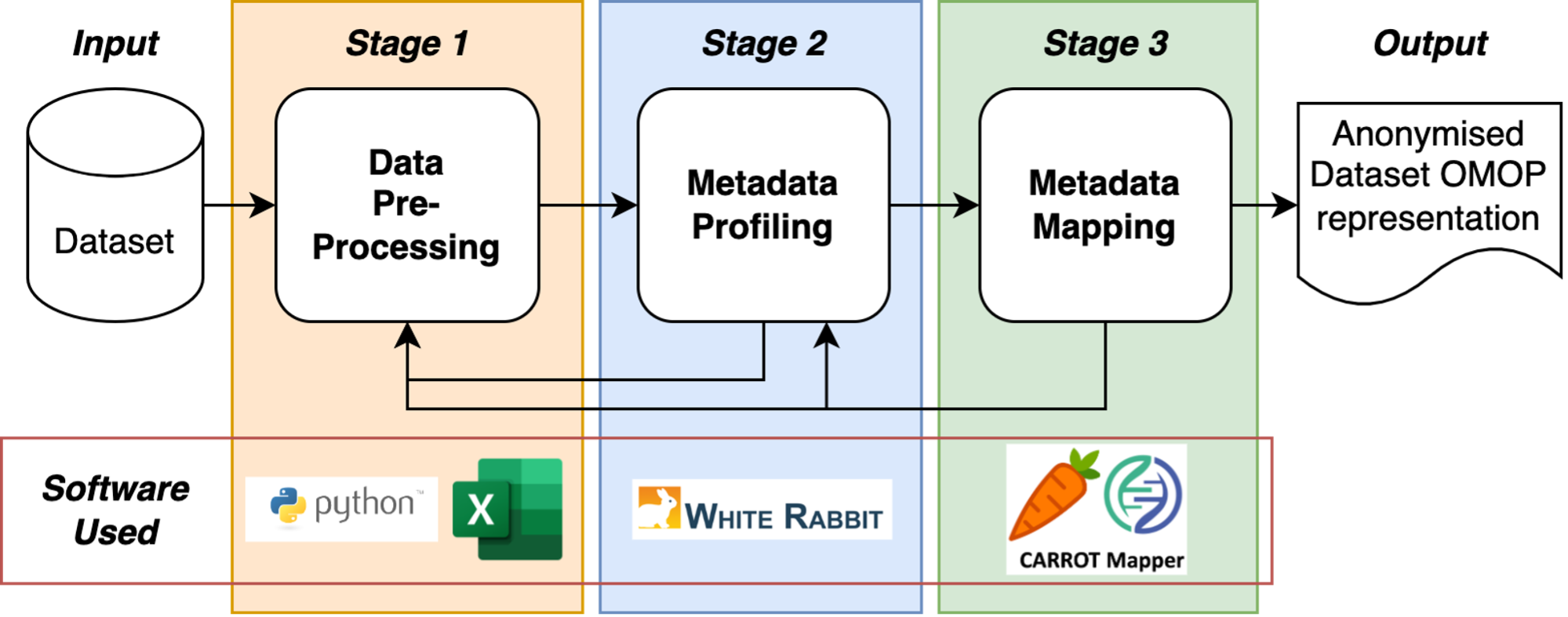
Figure 1: The stages of Health Informatics Data Engineering.
Data Engineering Overview
Figure 2 portrays the high-level view of the 3 stages and the main processes they are comprised of:
- Stage 1: Data Pre-processing
The pre-processing stage focuses on the ‘important attributes’ of a dataset that need to be mapped to the OMOP Common Data Model for discoverability.
A meeting with the dataset owner is necessary to understand the context and select the key data attributes to be made discoverable. From this a subset of the data is then anonymised by the data owner and refactored (reorganised), producing a pre-processed dataset. This is then reviewed by the data owner to determine if it is fit for purpose and is fully anonymised.
The output of Stage 1 is an anonymised dataset ready for profiling together with a formatted data dictionary.
- Stage 2: Metadata Profiling
The metadata profiling stage assesses the structure of a pre-processed dataset.
The pre-processed dataset is loaded into the OHDSI WhiteRabbit tool, this produces a statsitical profile of the data called a ‘Scan Report’ that further de-identifies data by removing the relationships between data attributes. This is then checked to ensure that the data has been rigorously anonymised.
The output of Stage 2 is a Verified Scan Report spreadsheet.
- Stage 3: Metadata Mapping
The metadata mapping stage takes the dataset attributes and maps them to OHDSI OMOP Common Data Model for discoverability.
Individual dataset attributes are mapped to ‘Standard’ OMOP terms (e.g., these could be from SNOMED CT, RxNORM or LOINC controlled international standardised healthcare vocabularies), this process creates rules. The resultant rule set is then reviewed for correctness and completeness.
The output of Stage 3 is a mapped rule set in a JSON format.
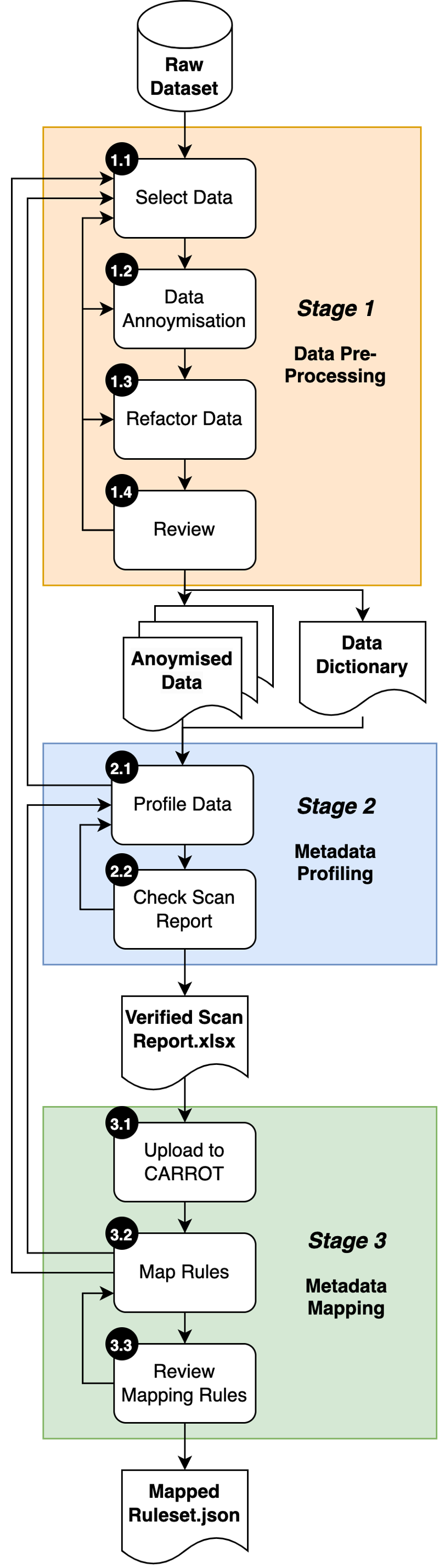
Figure 2: Data Engineering Overview
Reference
- Motwani, R., & Xu, Y. (2007). Efficient Algorithms for Masking and Finding Quasi-Identifiers. Available at: https://theory.stanford.edu/~xuying/papers/quasi_vldb1.pdf
- Wilkinson, M., Dumontier, M., Aalbersberg, I. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3, 160018 (2016). Available at: https://www.nature.com/articles/sdata201618
- OHDSI: https://www.ohdsi.org/
- WhiteRabbit: https://www.ohdsi.org/analy.c-tools/whiterabbit-for-etl-design/
- The FAIR principles explained: https://www.youtube.com/watch?v=5OeCrQE3HhE
- Information Commissioner’s Office - What is Personal Data: https://ico.org.uk/for-organisations/uk-gdpr-guidance-and-resources/personal-information-what-is-it/what-is-personal-data/what-is-personal-data/
- Devaux, E. (2020). How “anonymous” is anonymized data? Available at: https://medium.com/sta.ce/data-privacy-and-protection-techniques-db42576d8b3f#
Bibliography
- FAIR Principles: https://www.go-fair.org/fair-principles/
- Pseudonymization vs anonymization: differences under the GDPR: https://www.statice.ai/post/pseudonymization-vs-anonymization
- How “anonymous” is anonymized data?: https://www.statice.ai/post/how-anonymous-is-anonymized-data
- Lubarsky, B (2007). Re-identification of “anonymized data”. https://georgetownlawtechreview.org/wp-content/uploads/2017/04/Lubarsky-1-GEO.-L.TECH.-REV.-202.pdf